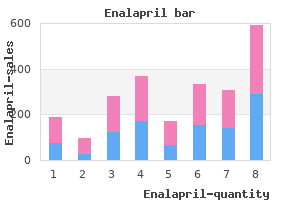
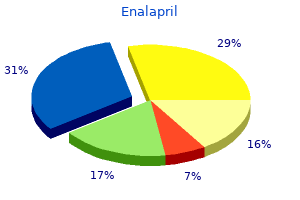
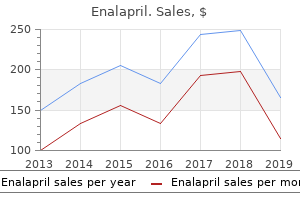
Enalapril"Buy discount enalapril 5 mg on-line, heart attack blood test". By: U. Kor-Shach, M.A., M.D. Program Director, Dartmouth College Geisel School of Medicine The frequency of aa types is determined by dividing the number of nontasters (37) by the total number of individuals (125) blood pressure 88 over 60 buy 10mg enalapril overnight delivery. Know- B-39 ing the frequencies of all the genotypes, one can calculate p as the sum of 0. Since the frequencies of all the genotypes are known, one can calculate p as the sum of 0. Notice that there are more heterozygotes and fewer homozygotes (especially the ss types) than predicted in the population. For each of these values, one merely takes the square root to determine q, then computes p, and then "plugs" the values into the 2pq expression. Because a dominant lethal gene is highly selected against, it Solutions to Problems and Discussion Questions 2. Your essay should include a discussion of the original sources of variation coming from mutation and that migration can cause gene frequencies to change in a population if the immigrants have different gene frequencies compared to the host population. You should also describe selection as resulting from the biased passage of gametes and offspring to the next generation. The classification of organisms into different species is based on evidence (morphological, genetic, ecological, etc. That is, there must be evidence that gene flow does not occur among the groups being called different species. Indeed, classification above the species level is somewhat arbitrary and based on traditions that is unlikely that it will exist at too high a frequency, if at all. However, if the gene shows incomplete penetrance or late age of onset (after reproductive age), it may remain in a population. The frequency of an allele is determined by a number of factors, including the fitness it confers, mutation rate, and input from migration. With one exception (the final amino acid in the sequences), are dominant-unless there is very strong selection for the allele. The distribution of a gene among individuals is determined by mating (population size, inbreeding, etc. This confusion often stems from the 1:2:1 (or 3:1) ratio seen in Mendelian crosses. The presence of selectively neutral alleles and genetic adaptations to varied environments contribute significantly to genetic variation in natural populations. Somatic gene therapy, like any therapy, allows some individuals to live more normal lives than those not receiving therapy. As such, the ability of these individuals to contribute to the gene pool increases the likelihood that less-fit alleles will enter and be maintained in the gene pool. This is a normal consequence of therapy, genetic or not, and in the face of disease control and prevention, societies have generally accepted this consequence. Germ-line therapy could, if successful, lead to limited, isolated, and infrequent removal of an allele from a gene lineage. However, given the present state of the science, its impact on the course of human evolution will be diluted and negated by a host of other factors that afflict humankind. Prezygotic mechanisms are more efficient because they occur before resources are expended in the processes of mating. In small populations, large fluctuations in gene frequency occur because random gametic sampling may not include all the genetic variation in the parents. Two factors can cause the extinction of a particular mutation in small populations. First, sampling error may allow fixation of one form and the elimination of others. If an advantageous mutation occurs, it must be included in the next replicative round in order to be maintained in subsequent generations.
Although this error rate may seem very low heart attack 40 year old male order line enalapril, it still allows about 600,000 errors in the human genome sequence. Celera sequenced certain areas of the genome more than 35 times when compiling the genome. Yet even at the time of "completion" there were still some 350 gaps in the sequence that continued to be worked on. Recall that the number of genes had originally been estimated to be about 100,000, based in part on a prediction that human cells produce about 100,000 proteins. At least half of the genes show sequence similarity to genes shared by many other organisms, and as you will learn in Section 21. There is still no consensus among scientists worldwide about the exact number of human genes. One reason is that it is unclear whether or not many of the presumed genes produce functional proteins. Currently, annotation predicts that the human genome encodes approximately 21,000 proteins. The number of genes is much lower than the number of predicted proteins in part because many genes code for multiple proteins through alternative splicing. Initial estimates suggested that over 50 percent of human genes undergo alternative splicing to produce multiple transcripts and multiple proteins. Clearly, alternative splicing produces an incredible diversity of proteins beyond simple predictions based on the number of genes in the human genome. After the draft sequence of the human genome was completed, it initially appeared that most genetic variations between individuals (the 0. Determining human gene functions, deciphering complexities of geneexpression regulation and gene interaction, and uncovering the relationships between human genes and phenotypes are among the many ongoing challenges for genome scientists. Accessing the Human Genome Project on the Internet It is now possible to access databases and other sites on the Internet that display maps for all human chromosomes. You may already have used Map Viewer for the Exploring Genomics exercises earlier in the text (see Chapters 5 and 12). Maps such as this depict genes thought to be involved in human genetic disease conditions. To the right of the ideogram is a column showing the contigs (arranged vertically) that were aligned to sequence this chromosome. The UniGene column displays a histogram representation of gene density on chromosome 12. Finally, gene symbols, loci, and gene names (by description) are provided for selected genes; in this figure only 20 genes are identified. When accessing these maps on the Internet, one can magnify, or zoom in on, each region of the chromosome, revealing all genes mapped to a particular area. Thus, extensive maps have been developed for genes implicated in human disease conditions. This provides functional classifications of genes and encoded proteins using data from functional genomics, experimental approaches, and other analyses. We will discuss aspects of a cancer genome project (Cancer Genome Atlas Project) later in the text (see Chapter 24). Two programs funded by the National Institutes of Health challenged scientists to develop sequencing technologies to complete a human genome for $1000 by 2014 (see the Genetics, Technology, and Society essay in Chapter 22). As you learned earlier in the text (see Chapter 20), several companies have now developed technology that can sequence a genome for less than $1000. James Watson, a woman with acute myeloid leukemia, a Yoruba male from Nigeria and the first Asian genome J. Craig Venter diploid genome Billions of base pairs 200 150 Automated Sanger Sequencing: At the peak of this technique, a single machine could produce hundreds of thousands of base pairs in a single run. Others, such as Pacific Biosciences, and Ion Torrent, can read from longer molecules as they pass through a pore. Sequencing costs have steadily declined since 2000 due to innovations in sequencing technology. As a result, notice that the number of individual genomes sequenced has dramatically increased. As you will learn later in in the text (see Chapter 22), having somebody such as a geneticist analyze your personal genome data and consider how genome variations may affect your health may be expensive.
Nod1 and nod2 are expressed in human and murine renal tubular epithelial cells and participate in renal ischemia reperfusion injury blood pressure monitor reviews purchase enalapril 10mg fast delivery. Phenotyping of Nod1/2 double deficient mice and characterization of Nod1/2 in systemic inflammation and associated renal disease. Role of the nucleotide-binding domain-like receptor protein 3 inflammasome in acute kidney injury. Mangiferin attenuate sepsis-induced acute kidney injury via antioxidant and anti-inflammatory effects. Discuss the role novel biomarkers can play in providing information on kidney damage and the latest markers to assess kidney stress. A week was selected arbitrarily as a time course between the presumed exposure and the clinical manifestations of either oliguria or azotemia. Of course a week is a long time for oliguria to occur, but serum creatinine may well not reach steady state for longer than a week. Furthermore, because it is often difficult (if not impossible) to determine the precise start of the injury, it is unclear when the clock starts. For example, in a patient admitted on Friday with unstable angina who then has three daily serum creatinine measures, all essentially the same, before undergoing cardiac surgery on Monday, there is no need to have a historical baseline to evaluate the serum creatinine on postop day 1. By contrast, consider a patient who has a 2-day history of fever and cough and an elevated creatinine. Perhaps the creatinine continues to increase after admission and does not recover. For many patients, the best reference creatinine will not be the admission value, because it is likely already abnormal (unless the patient presented only with oliguria). Therefore a baseline creatinine obtained before the current illness but still recent would be ideal. As such we are left with deciding between various less ideal baseline values or no value at all. Various studies have shown that even an old baseline (up to 1 year prior) is better than nothing. Because a normal creatinine may vary by more than twofold based on demographics (especially age, race, and sex), it is not appropriate to use a single normal value for all patients. Recovery by hospital discharge still was achieved by 61% of those relapsing, but those who did not recover outcomes at 1 year were markedly worse. The criteria for acute kidney injury are based on acute changes in serum creatinine and urine output. Baseline renal function also is based on clinical judgment and is best determined by prior serum creatinine measurements; when none are available estimating equations can be used with caution. Novel biomarkers can provide information on kidney damage, and the latest markers can assess kidney stress. Transient and persistent acute kidney injury and the risk of hospital mortality in critically ill patients: results of a multicenter cohort study. Novel urinary tubular injury markers reveal an evidence of underlying kidney injury in children with reduced left ventricular systolic function: a pilot study. Recognize acute kidney injury is associated with significant long-term clinical outcomes. Because of these limitations, there is a need for biomarkers to assist in detection of ongoing injury, repair, and recovery of renal function. A definition for renal recovery is required first before uniform detection of persistent renal dysfunction and categorization can be performed. Some studies defined complete recovery as a return of serum creatinine to within 20% of baseline, whereas partial recovery was anything short of continued dialysis dependence. The degree of recovery and timing of recovery are associated with important clinical outcomes,16,55 so definitions should incorporate this and be tested regarding ability to assess future risk. Development of proteinuria or albuminuria may be useful to define continued renal dysfunction, but baseline and follow-up measures are often unavailable for analysis in observational studies. One estimate for the developed nations (United States, Canada, Western Europe, and Australia) is 1. Discharge planning and systematic postdischarge followup (for all hospitalizations or for designated at-risk individuals) show promise in reduction of hospital readmissions and possibly mortality67 as well as improved access for vulnerable populations. Further refinement of this inclusion policy may be possible through associational data.
Syndromes
We fail to reject the null hypothesis and are confident that the observed values do not differ significantly from the expected values blood pressure purchase 10 mg enalapril with visa. When homologous chromosomes separate from each other at anaphase I, alleles will go to opposite poles of the meiotic apparatus. Different gene pairs on the same homologous pair of chromosomes (if far apart) or on nonhomologous chromosomes will separate independently from each other during meiosis. In Cross 2, a typical 3:1 Mendelian ratio is observed, which indicates that two heterozygotes were crossed: Ww * Ww 14. We would therefore say that there is a "good fit" between the observed and expected values. The only difference is that the recessive genes are coming from both parents, rather than from one parent only as in part (a). When you have genes on the autosomes (not X-linked), independent assortment, complete dominance, and no gene interaction (see later) in a cross involving double heterozygotes, the offspring ratio will be 9:3:3:1. Even though this cross involves two gene pairs, it will give a "monohybrid" type of ratio because one of the gene pairs is homozygous (body color) and one gene pair is heterozygous (wing length). As the critical p value is increased, it takes a smaller x2 value to cause rejection of the null hypothesis. It would take less difference between the expected and observed values to reject the null hypothesis; therefore, the stringency of failing to reject the null hypothesis is increased. Under that circumstance, there is a 25 percent chance that each of their children would be affected. The probability that two children of heterozygous parents would be affected would be 0. In the first cross, notice that a 3: 1 ratio exists for the spiny to smooth phenotypes, leading to the hypothesis that the spiny allele is dominant to smooth. Apply the same reasoning to the second cross, where there is a 3: 1 ratio of purple to white. The purple, spiny F1 would support the hypothesis that purple is dominant to white and spiny is dominant to smooth. In the F2, a 9: 3: 3: 1 ratio would not only support the above hypothesis, but also indicate the independent inheritance and expression of the two traits. Also, there are only three possibilities: both are heterozygous, neither is heterozygous, and at least one is heterozygous. You have already calculated the first two probabilities; the last is simply 1 - (1/12 + 6/12) = 5/12. One would reject the null hypothesis and assume a significant difference between the observed and expected values. In fact, most statisticians recommend that the expected values in each class should not be less than 10. Crosses: ckck ckck ckcd ckcd ckca ckca * * * * * * cdcd cdca cdcd cdca cdcd cdca all sepia all sepia 1/2 sepia; 1/2 cream 1/2 sepia; 1/2 cream 1/2 sepia; 1/2 cream 1/2 sepia; 1/4 cream; 1/4 albino (d) Parents: sepia * cream Because the sepia parent had a full color parent and an albino parent (Cck * caca), it must be ckca. The cream parent had two full color parents that could be Ccd or Cca; therefore, it could be cdcd or cdca. Crosses: ckca * cdcd ckca * cdca b = red 1/2 sepia; 1/2 cream 1/2 sepia; 1/4 cream; 1/4 albino 4. C chC ch = chestnut C cC c = cremello C chC c = palomino (b) the F1 resulting from matings between cremello and chestnut horses would be expected to be all palomino. The F2 would be expected to fall in a 1: 2: 1 ratio as in the third cross in part (a). F2 offspring would have the following "simplified" genotypes with the corresponding phenotypes: A C = 9/16 (agouti) A cc = 3/16 (colorless because cc is epistatic to A) aaC = 3/16 (black) aacc = 1/16 (colorless because cc is epistatic to aa) the two colorless classes are phenotypically indistinguishable; therefore, the final ratio is 9: 3: 4. Half of the pigmented offspring are black and half are agouti; therefore, the female must have been Aa. Discount enalapril 10mg mastercard. Ano Ang Resistant Hypertension ?.
|