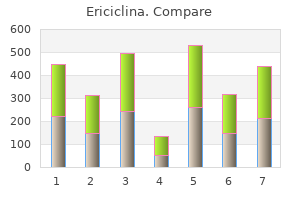
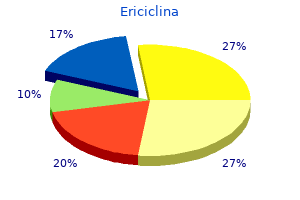
Ericiclina"Discount 100 mg ericiclina amex, virus 7912". By: R. Orknarok, M.B.A., M.B.B.S., M.H.S. Professor, Texas Tech University Health Sciences Center School of Medicine The a cell and the a cell each encodes cell-type-specific regulators: a cells make the regulatory protein a1 oral antibiotics for sinus infection buy ericiclina 500 mg without a prescription, and a cells make the proteins a1 and a2. A fourth regulatory protein, called Mcm1, is also involved in regulating the mating-type-specific genes (and many other genes) and is present in both cell types. In a cells, the a-specific genes are off because no activators are bound there, whereas the a-specific genes are on because Mcm1 is bound and activates those genes. In a cells, the a-specific genes are on because Mcm1 is bound upstream and activates them. At these genes, Mcm1 binds to a weak site and does so only when it binds cooperatively with a monomer of the protein a1. One ubiquitous regulator (Mcm1) and three cell-type-specific regulators (a1, a1, and a2) together regulate three classes of target genes. Transcriptional Regulation in Eukaryotes 681 perties of a2 ensure that a-specific genes are not expressed here: it covers the activating region of Mcm1, preventing that protein from activating; it also actively represses the genes. This is done as follows: the a-specific genes bind Mcm1 and a2, just as they do in a cells. Both the haploid cell types (a and a) express another class of genes called haploid-specific genes. These are switched off in the diploid cell by a2, which binds upstream of them as a heterodimer with the a1 protein. The molecular details of mating-type gene regulation are now known for other yeast species. In Box 19-4, Evolution of a Regulatory Circuit, we compare how a-specific and a-specific genes are regulated in S. The comparison reveals how a gene regulatory circuit can evolve, a topic we return to in subsequent chapters. But we also saw other ways they can work: they can bind to sites adjacent to promoters and, by interacting with polymerase bound there, inhibit the enzyme from initiating transcription. In eukaryotes, we see all of these except the first (ironically, the most common in bacteria). We also see another form of repression, perhaps the most common in eukaryotes, that works as follows. As with activators, repressors can recruit nucleosome modifiers, but in this case, the enzymes have effects opposite to those recruited by activators-they compact the chromatin or remove groups recognized by the transcriptional machinery. Therefore, for example, histone deacetylases repress transcription by removing acetyl groups from the tails of histones in S. Paradoxically, the histone deacetylase Rpd3 is also recruited to active genes to ensure transcription fidelity. Other enzymes add methyl groups to histone tails, and this frequently represses transcription, although in some cases it is associated with an actively transcribed gene (see Chapter 8). In a variation on this theme, a repressor can be a derivative of the same protein asthe activator but lack the activating region. Repression is likely the result of deacetylation of local nucleosomes (Tup1 recruits a deacetylase) and also perhaps of directly contacting and inhibiting the transcriptional machinery. First, Tup1 acts on nucleosomes either through recruiting histone deacetylases and/or by positioning a nucleosome at or near the transcription start site. Second, Tup1 interacts directly with the transcriptional machinery at the promoter and inhibits initiation. Thus, in a cells, the a-specific genes are expressed and the a-specific genes are not, whereas in a cells, the a-specific genes are expressed and the a-specific genes are not. Another class of genes-haploid-specific genes (hsgs)-is expressed in both of these cell types (a and a) but not in the diploid a/a cell. The products of the hsgs are required for mating-something a and a cells do, but that a/a cells (the product of the mating) cannot. Recent studies provide an illuminating example of how gene regulatory networks evolve. If expressed in terms of the divergence of conserved proteins, these two yeast are more divergent than fish and mammals. That nucleotide is then chosen as the nucleotide of choice at that position in the consensus; its relative frequency and the frequencies with which the other three nucleotides occur at each position are portrayed in the graph virus 1999 torrent discount ericiclina online. In that example, each individual promoter sequence had previously been identified, thus aligning the sequences is tri- vial. However, several regions of a chromosome are known to contain binding sites somewhere within their lengths. A computer algorithm is used that scans each of the sequences of these chromosomal regions, searching for a potential binding site common to them all. A comparison of the sequences bound reveals the consensus readily, because each of the fragments is very short. The correlation between promoter strength and sequence explains why promoters are so heterogeneous: some genes need to be expressed more highly than others, and the former are likely to have sequences closer to the consensus. The strength of the interaction between the discriminator and polymerase influences the stability of the complex between the enzyme and the promoter. The s Factor Mediates Binding of Polymerase to the Promoter the s70 factor can be divided in to four regions called s region 1 through s region 4. Two bases in the non-template strand are flipped out and inserted in to pockets within the s protein where they make favorable contacts that stabilize the unwound state of the promoter region. This helix makes contact with the two specific base pairs that constitute that element. Those regions of s factor that recognize specific regions of the promoter are indicated by arrows. The s subunit is positioned within the holoenzyme structure in such a way as to make feasible the recognition of various promoter elements. This rather large spacing of the protein domains is accommodated by the region between s regions 2 and 4, that is, by region 3-especially region 3. The next stage in initiation requires the enzyme to become more intimately engaged with the promoter, in the open complex. Isomerization is essentially irreversible and, once complete, typically guarantees that transcription will subsequently initiate (although regulation can still be imposed after this point in some cases). Formation of the closed complex, in contrast, is readily reversible: polymerase can as easily dissociate from the promoter as make the transition to the open complex. To picture the global structural changes within the polymerase that accompany isomerization, we need to examine the structure of the holoenzyme in more detail. The cut-away reveals the two flipped-out bases, A and T (yellow), in the binding pockets. Red arrows show how the flipped-out bases relate to the same nucleotides in the closed complex. The active site of the enzyme, which is made up of regions from both the b and b0 subunits, is found at the base of the pincers within the active center cleft. Buy ericiclina with american express. How to Avoid C dif: watch out for antibiotics! IDWEEK 2017/CDC: watch your dentist doc hospital.
For example virus protection free 100 mg ericiclina with amex, if binding of a ligand to an enzyme stabilizes a conformation in which the active site is blocked, the ligand will have turned off the activity of that enzyme. Conversely, ligand binding at a remote site might favor a conformation in which the active site is available to substrate and complementary to the transition state of the reaction; the ligand would then be an activator. This kind of regulation is known as allosteric regulation or allostery, because the structure of the ligand (its "steric" character) is different from (Greek allo-) the structure of any of the reactants. The Lac repressor (which inhibits expression of the bacterial gene encoding b-galactosidase, an enzyme that hydrolyzes b-galactosides such as lactose) is a good example of allosteric regulation in control of transcription. Even more complicated allosteric switches are possible, with multiple ligands and multiple binding sites. Allosteric regulation often involves quaternary-structure changes, as in the transition between the two dimer conformations of Lac repressor. The 20 L-amino acids specified by the genetic code include nine with nonpolar (hydrophobic) side chains, six with polar side chains that do not bear a charge at neutral pH, two with acidic side chains (negatively charged at neutral pH), and three with basic side chains (positively charged at neutral pH, or partially so in the case of histidine). Three amino acids have special conformational properties: glycine is nonchiral, with greater conformational freedom than the others; proline (technically, an imino acid) has a covalent bond between side chain and amide, restraining its conformational freedom; and cysteine, with a sulfhydryl group on its side chain, can undergo oxidation to form a disulfide bond with a second cysteine, crosslinking a folded polypeptide chain or two neighboring polypeptide chains. The reducing environment of a cell interior restricts disulfide-bond formation to oxidizing organelles and the extracellular milieu. Protein structure is traditionally described at four levels: primary (the sequence of amino acids in the polypeptide chain-the one level determined directly by the genetic code), secondary (local, repeated backbone conformations, stabilized by main-chain hydrogen bonds-principally a helices and b strands), tertiary (the folded, three-dimensional conformation of a polypeptide chain), and quaternary (association of folded polypeptide chains in a multisubunit assembly). At the tertiary level, polypeptide chains fold in to one or more independent domains, which would fold similarly even if excised from the rest of the protein. The structure of a domain can usefully be specified by the way in which its component secondary-structure elements (helices and strands) pack together in three dimensions. Linkers between domains of a multidomain polypeptide chain can be long and flexible or short and stiff. The aqueous environment and the diverse set of naturally occurring amino acids are together critical for the conformational stability of folded domains and of the interfaces between them that create quarternary structure. Nonpolar side chains cluster away from water in to the closely packed, hydrophobic core of a folded domain, and any sequestered hydrogen-bonding groups, which lose a hydrogen bond with water, must have a protein-derived partner. Secondary-structure elements satisfy the latter requirement for the main-chain amide and carbonyl groups, thus accounting for their importance in describing and classifying domain structures. The sequence of amino acids in a polypeptide chain specifies whether and how it will fold. This property allows the genetic code to determine not merely primary structure, but other levels as well, and hence to dictate protein function. The various noncovalent interactions within a correctly folded domain (and in extracellular domains, the covalent disulfide bonds) create a global free-energy minimum (conformation of greatest stability), so that the chain can reach its native conformation spontaneously. Changes in the environment of a protein, including post-translational modifications of one or more of its side chains or binding of ligands, may alter the position of this free-energy minimum and 144 Chapter 6 induce a conformational change. The array of amino acid side chains on the surface of a folded protein, and sometimes even in a segment of unfolded polypeptide chain, can also specify how it recognizes a protein or nucleic-acid partner or a small-molecule ligand. Proteins are thus the key agents of specific molecular recognition, both within a cell and between cells, as well as the specific catalysts of chemical reactions (enzymes). How does this interaction differ from the interactions that can occur between other amino acid side chains Give an example of two amino acid side chains that can interact with each other through an ionic bond at neutral pH. Amino acid substitutions are described as conservative when the amino acid in the mutated protein has chemical properties similar to those of the amino acid it has replaced. Explain what this statement means and why peptide bond formation is also referred to as a dehydration reaction. The oxygen-binding proteins hemoglobin and myoglobin differ in that hemoglobin functions as a tetramer in red blood cells, whereas myoglobin functions as a monomer in muscle cells. The globular structure of myoglobin and each hemoglobin monomer involves eight a-helical segments. Is it the primary, secondary, tertiary, or quaternary structure that differs most between these two proteins For the following amino acids, suggest whether they are more likely to be found buried or exposed in a stably folded For instructor-assigned tutorials and problems, go to MasteringBiology. For each interaction or bond below, state if the interaction or bond is disrupted by the urea treatment.
Often that is not good enough; sometimes antibiotics for dogs eye order 250mg ericiclina fast delivery, a gene must be completely shut off, on occasion permanently. The methyl group is added to the 50 position in the cytosine ring, generating 5-methylcytosine (see Chapter 4). This modification alone can disrupt binding of the transcription machinery and activators in some cases. These proteins, in turn, recruit complexes that remodel and modify local nucleosomes, switching off expression of the gene completely. Shown are two examples of genes controlled by imprinting-the mammalian Igf2 and H19 genes. As described in the text, in a given cell, the H19 gene is expressed only from the maternal chromosome, whereas Igf2 is expressed from the paternal chromosome. A signal released by one cell during development causes neighboring cells to switch on specific genes. These genes may have to remain switched on in those cells for many cell generations, even if the signal that induced them is present only fleetingly. The inheritance of gene expression patterns, in the absence of the initiating signal, is called epigenetic regulation. If a gene is controlled by an activator and that activator is only active in the presence of a given signal, then the gene will remain on only as long as the signal is present. Some States of Gene Expression Are Inherited through Cell Division Even When the Initiating Signal Is No Longer Present We have already encountered examples of gene regulation that can be inherited epigenetically. This state is associated with a specific pattern of gene expression and in particular with sustained expression of the l repressor protein (see Chapter 18. Lysogenic gene expression is established in an infected cell in response to poor growth conditions. Once established, however, the lysogenic state is maintained stably despite improvements in growth conditions: moving a lysogen in to rich growth medium does not lead to induction. Maintenance of the lysogenic state through cell division is thus an example of epigenetic regulation. In the second step, repressor synthesis is maintained by autoregulation: repressor activates expression of its own gene (see Chapter 18. In this way, when the lysogenic cell divides, each daughter cell inherits a copy of the dormant phage genome and some repressor protein. This repressor is sufficient to stimulate further repressor synthesis from the phage genome in each cell. Much of gene regulation during the development of muticellular organisms works in just this way. For the shutdown state to keep a gene off permanently, the methylation state must be inherited through cell division. The completely unmethylated sequence is not recognized by this enzyme and thus remains unmethylated. This condition is characterized by loss of language and motor skills in early childhood, microcephaly, seizures, stereotypical behaviors (such as repetitive handwringing), and intermitted hyperventilation. This exciting finding makes therapeutic intervention in humans more feasible, if still difficult. This protein, a growth factor, has roles in brain development and in synaptic changes associated with learning and memory. The broad array of symptoms-from cognitive impairment to unusual gait-suggests that there are probably several genes whose misexpression is required for the full disease. The syndrome is also associated with disrupted expression of imprinted genes on chromosome 11p15. As we have described, the Igf2 gene is usually expressed monoallelically; that is, only one of the two alleles (in this case, the paternal allele) is expressed as a result of imprinting of the other. Nucleosome modifications could in principle provide the basis for epigenetic inheritance, although no examples of this have yet been found.
|